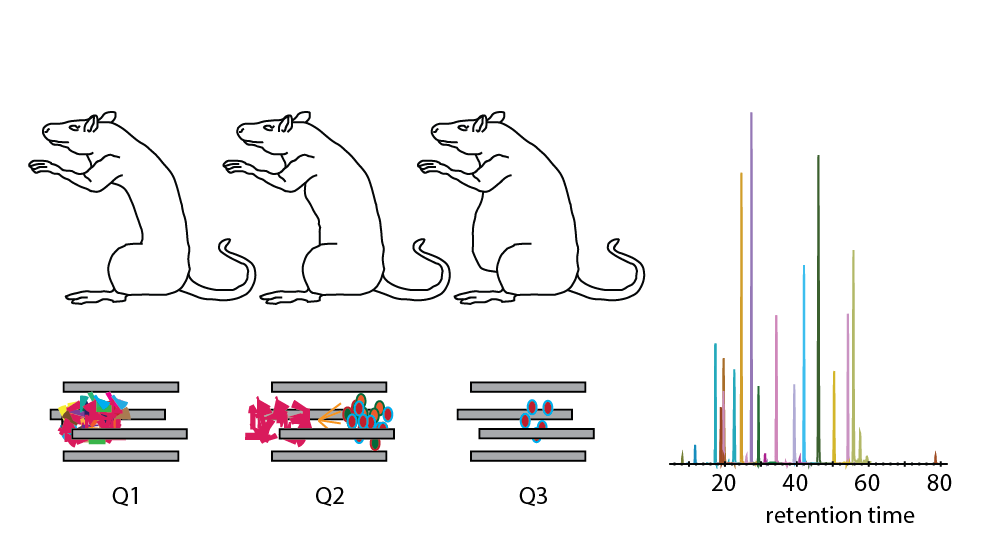
Understanding regulation and action of endogenous peptides, especially neuropeptides, which serve as inter- and intracellular signal transmitters, is key in understanding a variety of functional processes, such as energy balance, memory, circadian rhythm, drug addiction, etc. Therefore, accurate and reproducible quantification of these bioactive endogenous compounds is highly relevant. The biosynthesis of endogenous peptides, involving multiple possible trimming and modification events, hinders the de novo prediction of the active peptide sequences, making MS-based measurements very valuable in determining the actual active compounds. Here, we report an extended selected reaction monitoring (SRM)-based strategy to reproducibly and quantitatively monitor the abundances of a set of 15 endogenously occurring peptides from Rattus norvegicus hypothalamus. We demonstrate that SRM can be extended toward reproducible detection and quantification of peptides, bearing characteristics very different from tryptic peptides. We show that long peptide sequences, producing precursors with up to five and MS2 fragment ions with up to three charges, can be targeted by SRM on a triple quadrupole instrument. Using this approach to quantify endogenous peptide levels in hypothalami of animals subjected to different diets revealed several significant changes, most notably the significant upregulation of VGF-derived signaling peptide AQEE-30 upon high caloric feeding.
Schmidlin T, Boender AJ, Frese CK, Heck AJ, Adan RA, Altelaar AF.
Anal Chem. 2015 Oct 6;87(19):9966-9973